This document contains abridged sections from Discovering Statistics Using R and RStudio by Andy Field so there are some copyright considerations. You can use this material for teaching and non-profit activities but please do not meddle with it or claim it as your own work. See the full license terms at the bottom of the page.
Load packages
Remember to load the tidyverse:
library(tidyverse)
Load the data
Load the data from the discover package:
pupluv_tib <- discovr::puppy_love
If you want to read the file from the CSV and you have set up your project folder as I suggest in Chapter 1, then the code you would use is:
pupluv_tib <- here::here("data/puppy_love.csv") %>%
readr::read_csv() %>%
dplyr::mutate(
dose = forcats::as_factor(dose)
)
A good idea to check that the levels of dose are in the order Control, 15 minutes, 30 minutes by executing:
levels(pupluv_tib$dose)
## [1] "No puppies" "15 mins" "30 mins"
If they’re not in the correct order then:
pupluv_tib <- pupluv_tib %>%
dplyr::mutate(
dose = forcats::fct_relevel(dose, "No puppies", "15 mins", "30 mins")
)
Self-test
pupluv_tib %>%
dplyr::group_by(dose) %>%
dplyr::summarize(
dplyr::across(c(happiness, puppy_love), list(mean = mean, sd = sd), .names = "{col}_{fn}")
) %>%
dplyr::mutate(
dplyr::across(where(is.numeric), ~round(., 2))
)
| dose | happiness_mean | happiness_sd | puppy_love_mean | puppy_love_sd |
|---|
| No puppies | 3.22 | 1.79 | 3.44 | 2.07 |
| 15 mins | 4.88 | 1.46 | 3.12 | 1.73 |
| 30 mins | 4.85 | 2.12 | 2.00 | 1.63 |
pupluv_tib %>%
dplyr::summarize(
dplyr::across(c(happiness, puppy_love), list(mean = mean, sd = sd), .names = "{col}_{fn}")
) %>%
dplyr::mutate(
dplyr::across(where(is.numeric), ~round(., 2))
)
| happiness_mean | happiness_sd | puppy_love_mean | puppy_love_sd |
|---|
| 4.37 | 1.96 | 2.73 | 1.86 |
Self-test
pupluv_tib <- pupluv_tib %>%
dplyr::mutate(
none_vs_30 = ifelse(dose == "30 mins", 1, 0),
none_vs_15 = ifelse(dose == "15 mins", 1, 0)
)
pupluv_lm <- lm(happiness ~ none_vs_15 + none_vs_30 + puppy_love, data = pupluv_tib)
broom::glance(pupluv_lm)
| r.squared | adj.r.squared | sigma | statistic | p.value | df | logLik | AIC | BIC | deviance | df.residual | nobs |
|---|
| 0.2876499 | 0.2054557 | 1.743638 | 3.499636 | 0.0295449 | 3 | -57.10086 | 124.2017 | 131.2077 | 79.04712 | 26 | 30 |
broom::tidy(pupluv_lm, conf.int = TRUE) %>%
dplyr::mutate(
dplyr::across(where(is.numeric), ~round(., 3))
)
| term | estimate | std.error | statistic | p.value | conf.low | conf.high |
|---|
| (Intercept) | 1.789 | 0.867 | 2.063 | 0.049 | 0.007 | 3.572 |
| none_vs_15 | 1.786 | 0.849 | 2.102 | 0.045 | 0.040 | 3.532 |
| none_vs_30 | 2.225 | 0.803 | 2.771 | 0.010 | 0.575 | 3.875 |
| puppy_love | 0.416 | 0.187 | 2.227 | 0.035 | 0.032 | 0.800 |
Self-test
luvdose_lm <- lm(puppy_love ~ dose, data = pupluv_tib)
anova(luvdose_lm) %>%
parameters::model_parameters()
| Parameter | Sum_Squares | df | Mean_Square | F | p |
|---|
| dose | 12.76944 | 2 | 6.384722 | 1.979254 | 0.1577178 |
| Residuals | 87.09722 | 27 | 3.225823 | NA | NA |
Self-test
cov_first_lm <- lm(happiness ~ puppy_love + dose, data = pupluv_tib)
anova(cov_first_lm) %>%
parameters::model_parameters()
| Parameter | Sum_Squares | df | Mean_Square | F | p |
|---|
| puppy_love | 6.734357 | 1 | 6.734357 | 2.215050 | 0.1487004 |
| dose | 25.185194 | 2 | 12.592597 | 4.141929 | 0.0274465 |
| Residuals | 79.047116 | 26 | 3.040274 | NA | NA |
broom::tidy(cov_first_lm, conf.int = TRUE) %>%
dplyr::mutate(
dplyr::across(where(is.numeric), ~round(., 3))
)
| term | estimate | std.error | statistic | p.value | conf.low | conf.high |
|---|
| (Intercept) | 1.789 | 0.867 | 2.063 | 0.049 | 0.007 | 3.572 |
| puppy_love | 0.416 | 0.187 | 2.227 | 0.035 | 0.032 | 0.800 |
| dose15 mins | 1.786 | 0.849 | 2.102 | 0.045 | 0.040 | 3.532 |
| dose30 mins | 2.225 | 0.803 | 2.771 | 0.010 | 0.575 | 3.875 |
Self-test
dose_first_lm <- lm(happiness ~ dose + puppy_love, data = pupluv_tib)
anova(dose_first_lm) %>%
parameters::model_parameters()
| Parameter | Sum_Squares | df | Mean_Square | F | p |
|---|
| dose | 16.84380 | 2 | 8.421902 | 2.770113 | 0.0811729 |
| puppy_love | 15.07575 | 1 | 15.075748 | 4.958681 | 0.0348334 |
| Residuals | 79.04712 | 26 | 3.040274 | NA | NA |
broom::tidy(dose_first_lm, conf.int = TRUE) %>%
dplyr::mutate(
dplyr::across(where(is.numeric), ~round(., 3))
)
| term | estimate | std.error | statistic | p.value | conf.low | conf.high |
|---|
| (Intercept) | 1.789 | 0.867 | 2.063 | 0.049 | 0.007 | 3.572 |
| dose15 mins | 1.786 | 0.849 | 2.102 | 0.045 | 0.040 | 3.532 |
| dose30 mins | 2.225 | 0.803 | 2.771 | 0.010 | 0.575 | 3.875 |
| puppy_love | 0.416 | 0.187 | 2.227 | 0.035 | 0.032 | 0.800 |
Set contrasts
puppy_vs_none <- c(-2/3, 1/3, 1/3)
short_vs_long <- c(0, -1/2, 1/2)
contrasts(pupluv_tib$dose) <- cbind(puppy_vs_none, short_vs_long)
No covariate
pup_lm <- lm(happiness ~ dose, data = pupluv_tib)
anova(pup_lm)
## Analysis of Variance Table
##
## Response: happiness
## Df Sum Sq Mean Sq F value Pr(>F)
## dose 2 16.844 8.4219 2.4159 0.1083
## Residuals 27 94.123 3.4860
Type III sums of squares
pupluv_lm <- lm(happiness ~ puppy_love + dose, data = pupluv_tib)
car::Anova(pupluv_lm, type = 3)
## Anova Table (Type III tests)
##
## Response: happiness
## Sum Sq Df F value Pr(>F)
## (Intercept) 76.069 1 25.0205 3.342e-05 ***
## puppy_love 15.076 1 4.9587 0.03483 *
## dose 25.185 2 4.1419 0.02745 *
## Residuals 79.047 26
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Self test
dose_first_lm <- lm(happiness ~ dose + puppy_love, data = pupluv_tib)
car::Anova(cov_first_lm, type = 3)
## Anova Table (Type III tests)
##
## Response: happiness
## Sum Sq Df F value Pr(>F)
## (Intercept) 12.943 1 4.2572 0.04920 *
## puppy_love 15.076 1 4.9587 0.03483 *
## dose 25.185 2 4.1419 0.02745 *
## Residuals 79.047 26
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Adjusted means
modelbased::estimate_means(pupluv_lm, fixed = "puppy_love")
| dose | puppy_love | Mean | SE | CI_low | CI_high |
|---|
| No puppies | 2.733333 | 2.926370 | 0.5962045 | 1.700854 | 4.151886 |
| 15 mins | 2.733333 | 4.712050 | 0.6207971 | 3.435983 | 5.988117 |
| 30 mins | 2.733333 | 5.151251 | 0.5026323 | 4.118076 | 6.184427 |
Parameter estimates
broom::tidy(pupluv_lm, conf.int = TRUE) %>%
dplyr::mutate(
dplyr::across(where(is.numeric), ~round(., 3))
)
| term | estimate | std.error | statistic | p.value | conf.low | conf.high |
|---|
| (Intercept) | 3.126 | 0.625 | 5.002 | 0.000 | 1.841 | 4.411 |
| puppy_love | 0.416 | 0.187 | 2.227 | 0.035 | 0.032 | 0.800 |
| dosepuppy_vs_none | 2.005 | 0.720 | 2.785 | 0.010 | 0.525 | 3.485 |
| doseshort_vs_long | 0.439 | 0.811 | 0.541 | 0.593 | -1.228 | 2.107 |
Post hoc tests
modelbased::estimate_contrasts(pupluv_lm, fixed = "puppy_love")
| Level1 | Level2 | puppy_love | Difference | SE | CI_low | CI_high | t | df | p | Std_Difference |
|---|
| 15 mins | 30 mins | 2.733333 | -0.4392012 | 0.8112214 | -2.515070 | 1.6366676 | -0.5414073 | 26 | 0.5928363 | -0.2245258 |
| No puppies | 15 mins | 2.733333 | -1.7856801 | 0.8493553 | -3.959131 | 0.3877712 | -2.1023947 | 26 | 0.0907071 | -0.9128647 |
| No puppies | 30 mins | 2.733333 | -2.2248813 | 0.8028109 | -4.279228 | -0.1705345 | -2.7713641 | 26 | 0.0305250 | -1.1373905 |
Homogeneity of regression slopes
hors_lm <- lm(happiness ~ puppy_love*dose, data = pupluv_tib)
car::Anova(hors_lm, type = 3)
## Anova Table (Type III tests)
##
## Response: happiness
## Sum Sq Df F value Pr(>F)
## (Intercept) 53.542 1 21.9207 9.323e-05 ***
## puppy_love 17.182 1 7.0346 0.01395 *
## dose 36.558 2 7.4836 0.00298 **
## puppy_love:dose 20.427 2 4.1815 0.02767 *
## Residuals 58.621 24
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Alternatively, update the previous model:
hors_lm <- update(pupluv_lm, .~. + dose:puppy_love)
car::Anova(hors_lm, type = 3)
## Anova Table (Type III tests)
##
## Response: happiness
## Sum Sq Df F value Pr(>F)
## (Intercept) 53.542 1 21.9207 9.323e-05 ***
## puppy_love 17.182 1 7.0346 0.01395 *
## dose 36.558 2 7.4836 0.00298 **
## puppy_love:dose 20.427 2 4.1815 0.02767 *
## Residuals 58.621 24
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
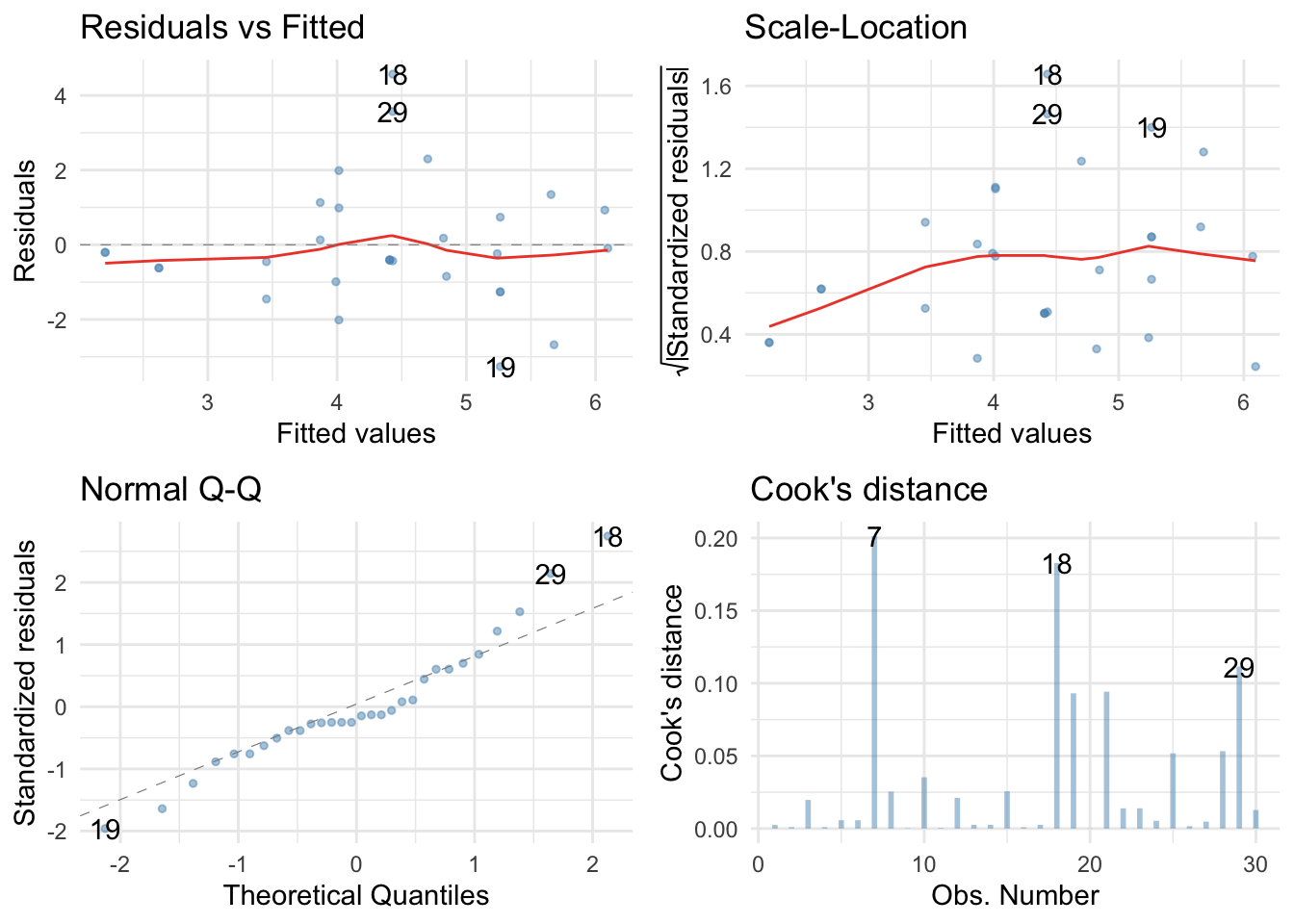
Residual plots:
ggplot2::autoplot(pupluv_lm,
which = c(1, 3, 2, 4),
colour = "#5c97bf",
smooth.colour = "#ef4836",
alpha = 0.5,
size = 1) +
theme_minimal()
robust model
Robust parameter estimates:
pupluv_rob <- robust::lmRob(happiness ~ puppy_love + dose, data = pupluv_tib)
summary(pupluv_rob)
##
## Call:
## robust::lmRob(formula = happiness ~ puppy_love + dose, data = pupluv_tib)
##
## Residuals:
## Min 1Q Median 3Q Max
## -2.34090 -0.34090 0.02081 0.89243 5.92569
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 2.1260 2.2616 0.940 0.356
## puppy_love 0.6333 0.5824 1.087 0.287
## dosepuppy_vs_none 1.6276 1.8641 0.873 0.391
## doseshort_vs_long -0.4549 2.8052 -0.162 0.872
##
## Residual standard error: 1.174 on 26 degrees of freedom
## Multiple R-Squared: 0.3901
##
## Test for Bias:
## statistic p-value
## M-estimate -5.868 1.0000
## LS-estimate 5.659 0.2261
Heteroscedasticity consistent standard errors:
parameters::model_parameters(pupluv_lm, robust = TRUE, vcov.type = "HC4", digits = 3)
| Parameter | Coefficient | SE | CI_low | CI_high | t | df_error | p |
|---|
| (Intercept) | 3.1260421 | 0.5920314 | 1.9091042 | 4.3429800 | 5.2801968 | 26 | 0.0000161 |
| puppy_love | 0.4160421 | 0.1985744 | 0.0078667 | 0.8242175 | 2.0951451 | 26 | 0.0460462 |
| dosepuppy_vs_none | 2.0052807 | 0.5157237 | 0.9451954 | 3.0653660 | 3.8882848 | 26 | 0.0006252 |
| doseshort_vs_long | 0.4392012 | 0.7399362 | -1.0817594 | 1.9601619 | 0.5935663 | 26 | 0.5579313 |
Bootstrap parameters
parameters::bootstrap_parameters(pupluv_lm)
| Parameter | Coefficient | CI_low | CI_high | p |
|---|
| (Intercept) | 3.1893353 | 2.1168995 | 4.5143321 | 0.0000000 |
| puppy_love | 0.3954619 | -0.0679983 | 0.7063347 | 0.0739261 |
| dosepuppy_vs_none | 1.9937821 | 0.8865644 | 3.0162401 | 0.0039960 |
| doseshort_vs_long | 0.3637506 | -0.8214466 | 1.8375369 | 0.5954046 |
Bayes factors
pupcov_bf <- BayesFactor::lmBF(formula = happiness ~ puppy_love, data = pupluv_tib, rscaleFixed = "medium", rscaleCont = "medium")
pupcov_bf
## Bayes factor analysis
## --------------
## [1] puppy_love : 0.6778239 ±0%
##
## Against denominator:
## Intercept only
## ---
## Bayes factor type: BFlinearModel, JZS
pup_bf <- BayesFactor::lmBF(formula = happiness ~ puppy_love + dose, data = pupluv_tib, rscaleCont = "medium", rscaleFixed = "medium")
pup_bf
## Bayes factor analysis
## --------------
## [1] puppy_love + dose : 1.431258 ±0.7%
##
## Against denominator:
## Intercept only
## ---
## Bayes factor type: BFlinearModel, JZS
pup_bf/pupcov_bf
## Bayes factor analysis
## --------------
## [1] puppy_love + dose : 2.111548 ±0.7%
##
## Against denominator:
## happiness ~ puppy_love
## ---
## Bayes factor type: BFlinearModel, JZS
Effect sizes
car::Anova(pupluv_lm, type = 3) %>%
effectsize::eta_squared(., ci = 0.95)
| Parameter | Eta_Sq_partial | CI | CI_low | CI_high |
|---|
| 2 | puppy_love | 0.1601709 | 0.95 | 0 | 0.4108051 |
| 3 | dose | 0.2416256 | 0.95 | 0 | 0.4733256 |
Without a pipe:
pupluv_aov <- car::Anova(pupluv_lm, type = 3)
effectsize::omega_squared(pupluv_aov, ci = 0.95)
| Parameter | Omega_Sq_partial | CI | CI_low | CI_high |
|---|
| 2 | puppy_love | 0.1165735 | 0.95 | -0.0370370 | 0.3795428 |
| 3 | dose | 0.1731860 | 0.95 | -0.0740741 | 0.4242189 |

