R code Chapter 10
This document contains abridged sections from Discovering Statistics Using R and RStudio by Andy Field so there are some copyright considerations. You can use this material for teaching and non-profit activities but please do not meddle with it or claim it as your own work. See the full license terms at the bottom of the page.
Load packages
Remember to load the tidyverse:
library(tidyverse)
Load the data
Remember to install the package for the book with library(discovr). After which you can load data into a tibble by executing:
name_of_tib <- discovr::name_of_data
For example, execute these lines to create the tibbles referred to in the chapter:
vids_tib <- discovr::video_games
infidelity_tib <- discovr::lambert_2012
newz_tib <- discovr::bronstein_2019
newz_md_tib <- discovr::bronstein_miss_2019
If you want to read the file from the CSV and you have set up your project folder as I suggest in Chapter 1, then the general format of the code you would use is:
tibble_name <- here::here("../data/name_of_file.csv") %>%
readr::read_csv() %>%
dplyr::mutate(
...
code to convert character variables to factors
...
)
In which you’d replace tibble_name with the name you want to assign to the tibble and change name_of_file.csv to the name of the file that you’re trying to load. You can use mutate to convert categorical variables to factors. For example, for the video_games data you would load it by executing:
library(here)
vids_tib <- here::here("data/video_games.csv") %>%
readr::read_csv()
Centering variables
A basic method
vids_tib <- discovr::video_games
vids_tib <- vids_tib %>%
dplyr::mutate(
caunts_cent = caunts - mean(caunts, na.rm = TRUE),
vid_game_cent = vid_game - mean(vid_game, na.rm = TRUE)
)
vids_tib
# Create a general function to do the centring
centre <- function(var){
var - mean(var, na.rm = TRUE)
}
# use the general function to centre multiple variables at once
vids_tib <- vids_tib %>%
dplyr::mutate(
dplyr::across(c(vid_game, caunts), list(cent = centre))
)
vids_tib
## # A tibble: 442 x 6
## id aggress vid_game caunts vid_game_cent caunts_cent
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 41xb 13 16 0 -5.84 -18.6
## 2 g4x6 38 12 0 -9.84 -18.6
## 3 31c4 30 32 0 10.2 -18.6
## 4 63ao 23 10 1 -11.8 -17.6
## 5 s17f 25 11 1 -10.8 -17.6
## 6 6qm9 46 29 1 7.16 -17.6
## 7 b74b 41 23 2 1.16 -16.6
## 8 w2l6 22 15 3 -6.84 -15.6
## 9 61i3 35 20 3 -1.84 -15.6
## 10 347s 23 20 3 -1.84 -15.6
## # … with 432 more rows
vids_tib <- vids_tib %>%
dplyr::mutate(
interaction = caunts_cent*vid_game_cent
)
vids_tib
Fitting the model
aggress_lm <- lm(aggress ~ caunts_cent + vid_game_cent + caunts_cent:vid_game_cent, data = vids_tib)
Or
aggress_lm <- lm(aggress ~ caunts_cent*vid_game_cent, data = vids_tib)
Viewing the model:
broom::glance(aggress_lm)
## # A tibble: 1 x 12
## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 0.377 0.373 9.98 88.5 9.16e-45 3 -1642. 3294. 3314.
## # … with 3 more variables: deviance <dbl>, df.residual <int>, nobs <int>
broom::tidy(aggress_lm, conf.int = TRUE)
## # A tibble: 4 x 7
## term estimate std.error statistic p.value conf.low conf.high
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 40.0 0.475 84.1 1.72e-272 39.0 40.9
## 2 caunts_cent 0.760 0.0495 15.4 6.12e- 43 0.663 0.857
## 3 vid_game_cent 0.170 0.0685 2.48 1.36e- 2 0.0350 0.304
## 4 caunts_cent:vid_gam… 0.0271 0.00698 3.88 1.22e- 4 0.0133 0.0408
Or, also round the digits
broom::tidy(aggress_lm, conf.int = TRUE) %>%
dplyr::mutate(
dplyr::across(where(is.numeric), ~round(., 3))
)
## # A tibble: 4 x 7
## term estimate std.error statistic p.value conf.low conf.high
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 40.0 0.475 84.1 0 39.0 40.9
## 2 caunts_cent 0.76 0.049 15.4 0 0.663 0.857
## 3 vid_game_cent 0.17 0.068 2.48 0.014 0.035 0.304
## 4 caunts_cent:vid_game_… 0.027 0.007 3.88 0 0.013 0.041
Fit a robust model:
parameters::model_parameters(aggress_lm, robust = TRUE, vcov.type = "HC4", digits = 3)
## Parameter | Coefficient | SE | 95% CI | t | df | p
## ------------------------------------------------------------------------------------------
## (Intercept) | 39.967 | 0.475 | [39.03, 40.90] | 84.136 | 438 | < .001
## caunts_cent | 0.760 | 0.047 | [ 0.67, 0.85] | 16.304 | 438 | < .001
## vid_game_cent | 0.170 | 0.076 | [ 0.02, 0.32] | 2.234 | 438 | 0.026
## caunts_cent * vid_game_cent | 0.027 | 0.007 | [ 0.01, 0.04] | 3.705 | 438 | < .001
Simple slopes and Johnson-Neyman interval
interactions::sim_slopes(
aggress_lm,
pred = vid_game_cent,
modx = caunts_cent,
jnplot = TRUE,
robust = TRUE,
confint = TRUE
)
## JOHNSON-NEYMAN INTERVAL
##
## When caunts_cent is OUTSIDE the interval [-17.10, -0.72], the slope of
## vid_game_cent is p < .05.
##
## Note: The range of observed values of caunts_cent is [-18.60, 24.40]
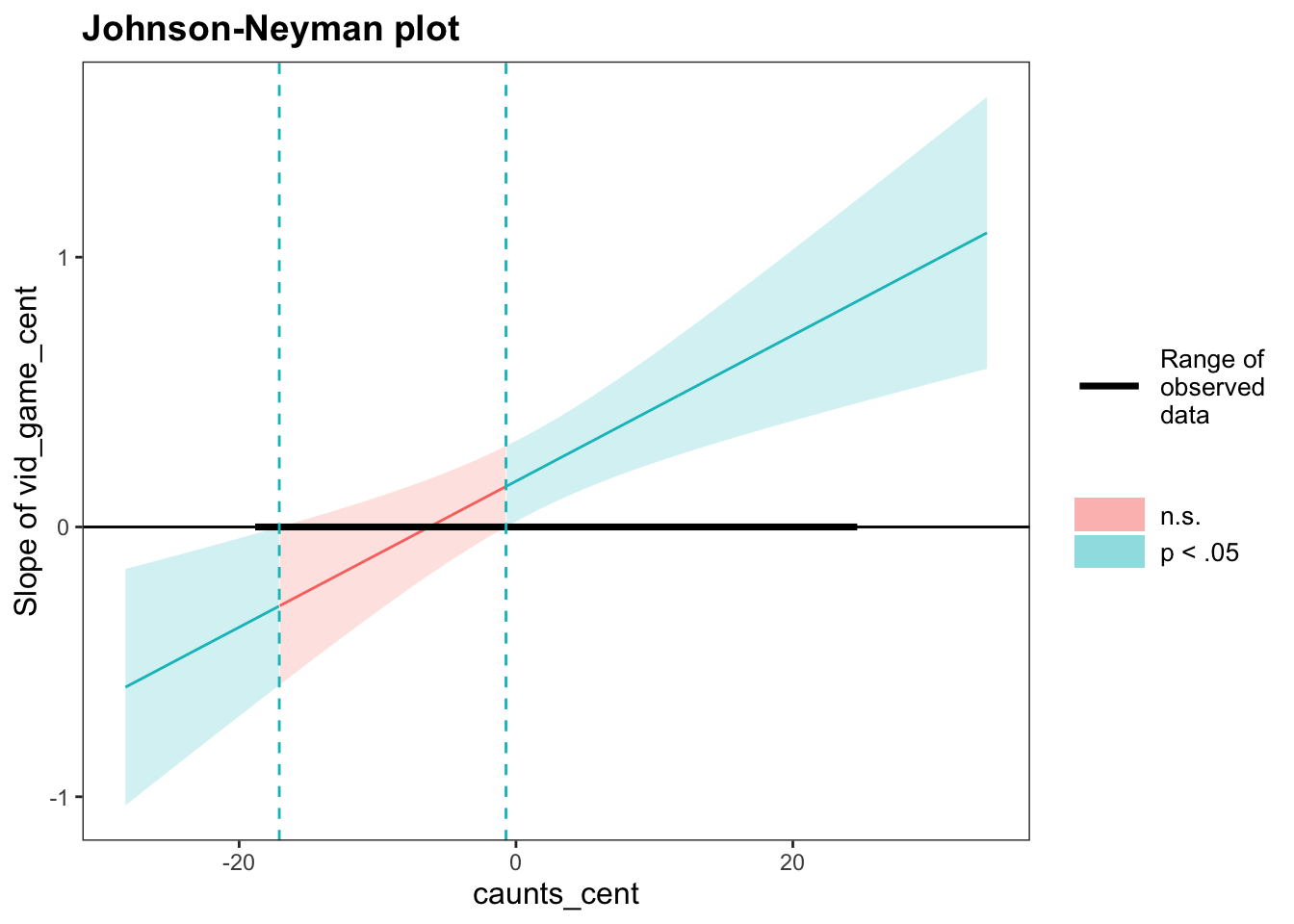
## SIMPLE SLOPES ANALYSIS
##
## Slope of vid_game_cent when caunts_cent = -9.62 (- 1 SD):
##
## Est. S.E. 2.5% 97.5% t val. p
## ------- ------ ------- ------- -------- ------
## -0.09 0.11 -0.30 0.12 -0.86 0.39
##
## Slope of vid_game_cent when caunts_cent = 0.00 (Mean):
##
## Est. S.E. 2.5% 97.5% t val. p
## ------ ------ ------ ------- -------- ------
## 0.17 0.08 0.02 0.32 2.23 0.03
##
## Slope of vid_game_cent when caunts_cent = 9.62 (+ 1 SD):
##
## Est. S.E. 2.5% 97.5% t val. p
## ------ ------ ------ ------- -------- ------
## 0.43 0.10 0.23 0.63 4.26 0.00
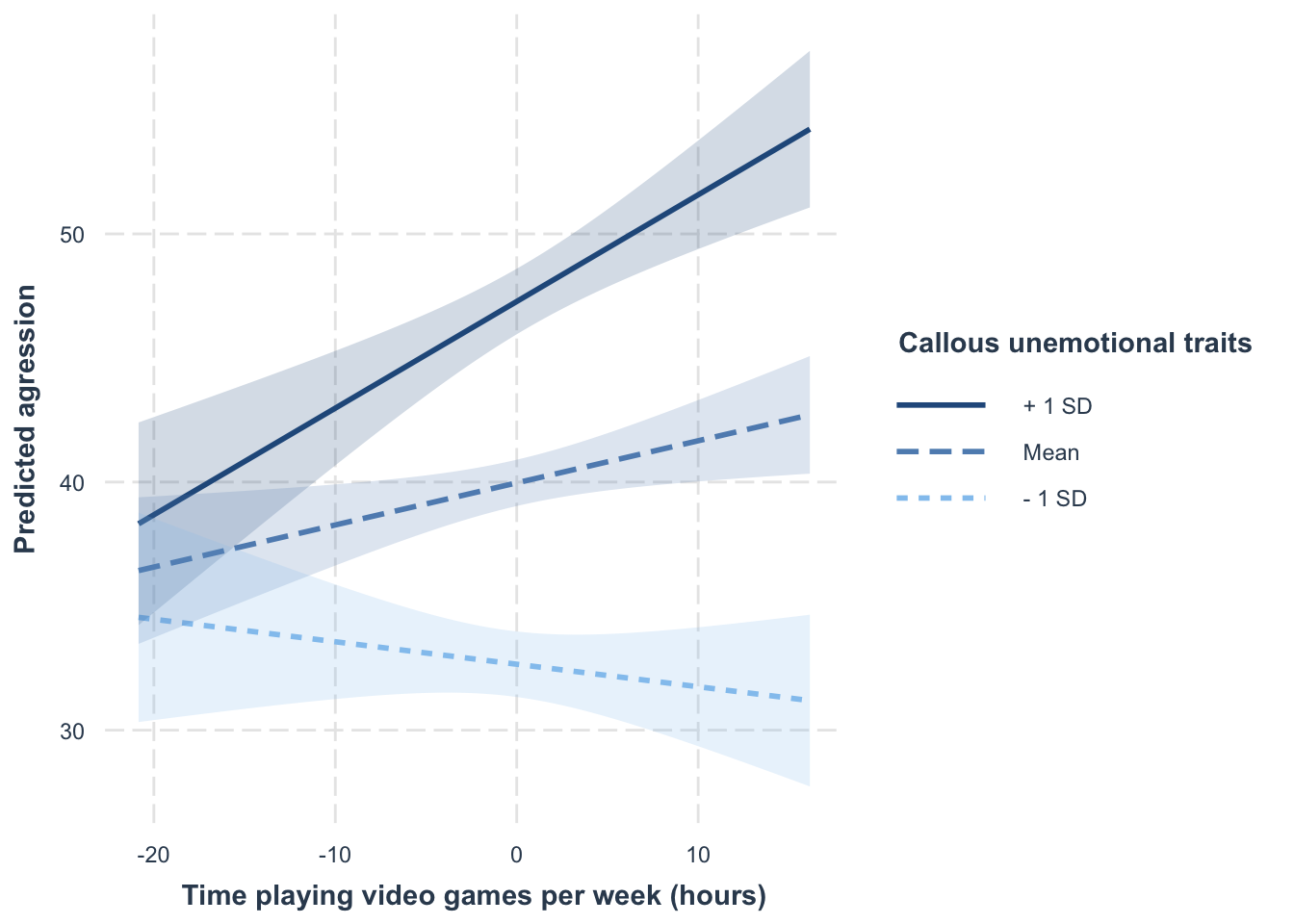
Simple slopes plot
interactions::interact_plot(
aggress_lm,
pred = vid_game_cent,
modx = caunts_cent,
interval = TRUE,
x.label = "Time playing video games per week (hours)",
y.label = "Predicted agression",
legend.main = "Callous unemotional traits"
)
Mediation
Self-test
m1 <- lm(phys_inf ~ ln_porn, data = infidelity_tib)
m2 <- lm(commit ~ ln_porn, data = infidelity_tib)
m3 <- lm(phys_inf ~ ln_porn + commit, data = infidelity_tib)
broom::tidy(m1, conf.int = TRUE)
## # A tibble: 2 x 7
## term estimate std.error statistic p.value conf.low conf.high
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0.230 0.0511 4.51 0.0000103 0.130 0.331
## 2 ln_porn 0.587 0.200 2.93 0.00369 0.193 0.981
broom::tidy(m2, conf.int = TRUE)
## # A tibble: 2 x 7
## term estimate std.error statistic p.value conf.low conf.high
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 4.20 0.0545 77.2 6.12e-170 4.10 4.31
## 2 ln_porn -0.470 0.213 -2.21 2.84e- 2 -0.889 -0.0501
broom::tidy(m3, conf.int = TRUE) %>%
dplyr::mutate(
dplyr::across(where(is.numeric), ~round(., 3))
)
## # A tibble: 3 x 7
## term estimate std.error statistic p.value conf.low conf.high
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 1.37 0.252 5.44 0 0.874 1.87
## 2 ln_porn 0.457 0.195 2.35 0.02 0.074 0.841
## 3 commit -0.271 0.059 -4.61 0 -0.387 -0.155
The better method
Look at patterns of missing data
infidelity_tib %>%
dplyr::select(id, commit, ln_porn, phys_inf) %>%
mice::md.pattern()
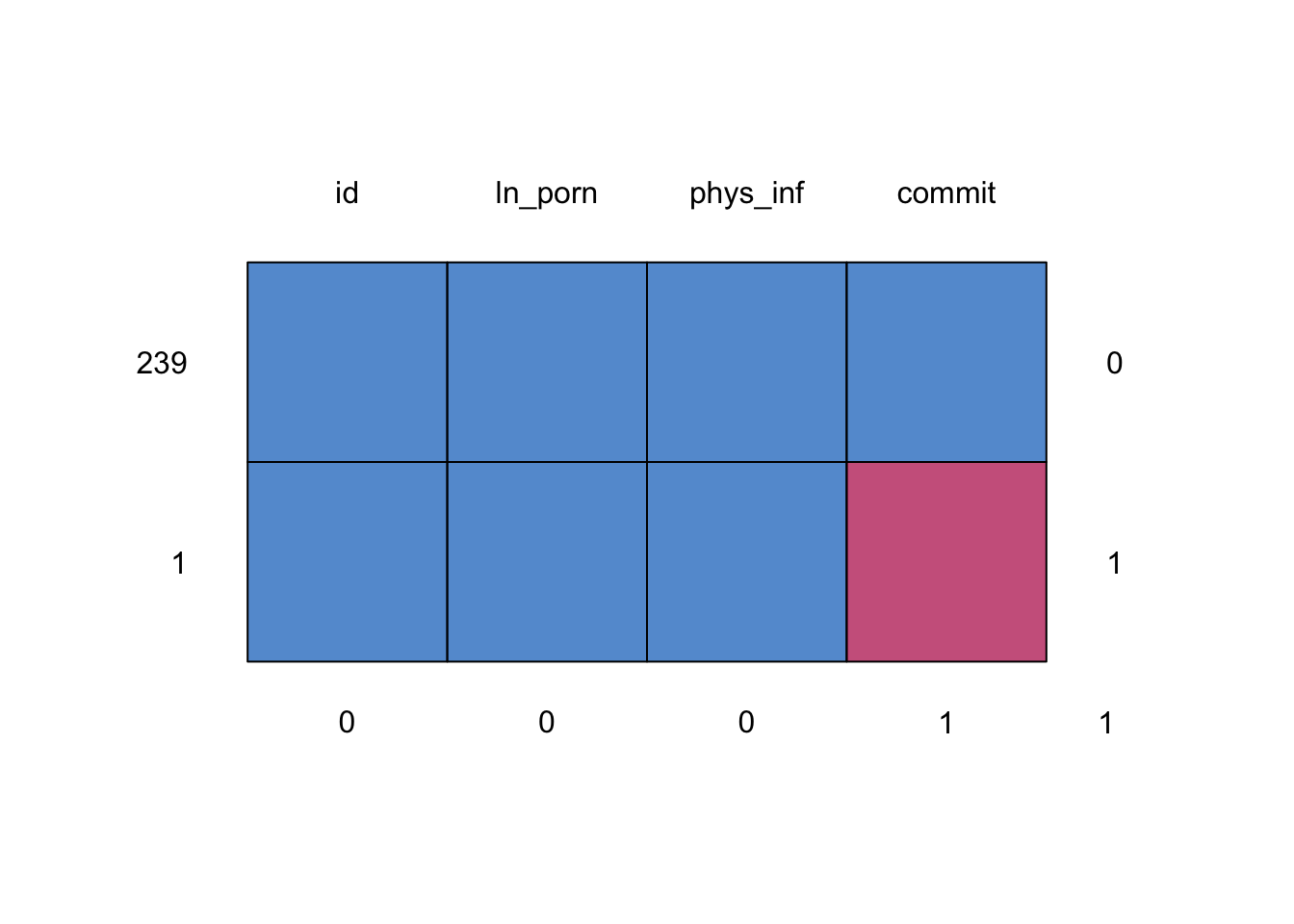
## id ln_porn phys_inf commit
## 239 1 1 1 1 0
## 1 1 1 1 0 1
## 0 0 0 1 1
infidelity_tib %>%
dplyr::select(id, commit, ln_porn, phys_inf) %>%
mice::ic()
## # A tibble: 1 x 4
## id commit ln_porn phys_inf
## <chr> <dbl> <dbl> <dbl>
## 1 1qx37 NA 0 0
infidelity_tib <- infidelity_tib %>%
dplyr::select(id, commit, ln_porn, phys_inf) %>%
na.omit()
Fit the model
infidelity_mod <- 'phys_inf ~ c*ln_porn + b*commit
commit ~ a*ln_porn
indirect_effect := a*b
total_effect := c + (a*b)
'
infidelity_fit <- lavaan::sem(infidelity_mod, data = infidelity_tib, missing = "FIML", estimator = "MLR")
broom::glance(infidelity_fit)
## # A tibble: 1 x 17
## agfi AIC BIC cfi chisq npar rmsea rmsea.conf.high srmr tli
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 1019. 1043. 1 0 7 0 0 4.90e-9 1
## # … with 7 more variables: converged <lgl>, estimator <chr>, ngroups <int>,
## # missing_method <chr>, nobs <int>, norig <int>, nexcluded <int>
broom::tidy(infidelity_fit, conf.int = TRUE) %>%
dplyr::mutate(
dplyr::across(where(is.numeric), ~round(., 3))
)
## # A tibble: 11 x 12
## term op label estimate std.error statistic p.value conf.low conf.high
## <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 "phy… ~ "c" 0.457 0.246 1.86 0.063 -0.024 0.939
## 2 "phy… ~ "b" -0.271 0.071 -3.81 0 -0.41 -0.132
## 3 "com… ~ "a" -0.47 0.229 -2.05 0.04 -0.918 -0.021
## 4 "phy… ~~ "" 0.432 0.054 7.98 0 0.326 0.539
## 5 "com… ~~ "" 0.531 0.05 10.7 0 0.433 0.629
## 6 "ln_… ~~ "" 0.049 0 NA NA 0.049 0.049
## 7 "phy… ~1 "" 1.37 0.316 4.34 0 0.751 1.99
## 8 "com… ~1 "" 4.20 0.053 79.5 0 4.10 4.31
## 9 "ln_… ~1 "" 0.126 0 NA NA 0.126 0.126
## 10 "ind… := "ind… 0.127 0.068 1.87 0.061 -0.006 0.26
## 11 "tot… := "tot… 0.585 0.244 2.40 0.016 0.107 1.06
## # … with 3 more variables: std.lv <dbl>, std.all <dbl>, std.nox <dbl>
Non-robust version for shits and giggles:
infidelity_nonrob <- lavaan::sem(infidelity_mod, data = infidelity_tib, missing = "FIML")
broom::tidy(infidelity_nonrob, conf.int = TRUE) %>%
dplyr::mutate(
dplyr::across(where(is.numeric), ~round(., 3))
)
## # A tibble: 11 x 12
## term op label estimate std.error statistic p.value conf.low conf.high
## <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 "phy… ~ "c" 0.457 0.193 2.37 0.018 0.078 0.836
## 2 "phy… ~ "b" -0.271 0.058 -4.64 0 -0.385 -0.157
## 3 "com… ~ "a" -0.47 0.212 -2.22 0.027 -0.885 -0.054
## 4 "phy… ~~ "" 0.432 0.04 10.9 0 0.355 0.51
## 5 "com… ~~ "" 0.531 0.049 10.9 0 0.436 0.626
## 6 "ln_… ~~ "" 0.049 0 NA NA 0.049 0.049
## 7 "phy… ~1 "" 1.37 0.25 5.48 0 0.88 1.86
## 8 "com… ~1 "" 4.20 0.054 77.5 0 4.10 4.31
## 9 "ln_… ~1 "" 0.126 0 NA NA 0.126 0.126
## 10 "ind… := "ind… 0.127 0.064 2.00 0.046 0.002 0.252
## 11 "tot… := "tot… 0.585 0.2 2.92 0.003 0.193 0.976
## # … with 3 more variables: std.lv <dbl>, std.all <dbl>, std.nox <dbl>
Standardized solution:
lavaan::standardizedsolution(infidelity_fit)
## lhs op rhs est.std se z pvalue ci.lower ci.upper
## 1 phys_inf ~ ln_porn 0.145 0.077 1.894 0.058 -0.005 0.296
## 2 phys_inf ~ commit -0.285 0.075 -3.814 0.000 -0.432 -0.139
## 3 commit ~ ln_porn -0.142 0.069 -2.069 0.039 -0.276 -0.007
## 4 phys_inf ~~ phys_inf 0.886 0.048 18.605 0.000 0.792 0.979
## 5 commit ~~ commit 0.980 0.019 50.415 0.000 0.942 1.018
## 6 ln_porn ~~ ln_porn 1.000 0.000 NA NA 1.000 1.000
## 7 phys_inf ~1 1.961 0.412 4.764 0.000 1.154 2.768
## 8 commit ~1 5.709 0.301 18.968 0.000 5.119 6.299
## 9 ln_porn ~1 0.569 0.000 NA NA 0.569 0.569
## 10 indirect_effect := a*b 0.040 0.021 1.902 0.057 -0.001 0.082
## 11 total_effect := c+(a*b) 0.186 0.075 2.466 0.014 0.038 0.334
Two mediators
newz_mod <- 'fake_newz ~ c*delusionz + b1*thinkz_open + b2*thinkz_anal
thinkz_open ~ a1*delusionz
thinkz_anal ~ a2*delusionz
indirect_open := a1*b1
indirect_anal := a2*b2
total_effect := c + (a1*b1) + (a2*b2)
thinkz_open ~~ thinkz_anal
'
newz_fit <- lavaan::sem(newz_mod, data = newz_tib, se = "bootstrap")
broom::tidy(newz_fit, conf.int = TRUE) %>%
dplyr::mutate(
dplyr::across(where(is.numeric), ~round(., 3))
)
## # A tibble: 13 x 12
## term op label estimate std.error statistic p.value conf.low conf.high
## <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 fake… ~ "c" 0.153 0.038 4.06 0 0.077 0.226
## 2 fake… ~ "b1" -0.171 0.035 -4.93 0 -0.239 -0.099
## 3 fake… ~ "b2" -0.141 0.035 -4.02 0 -0.209 -0.069
## 4 thin… ~ "a1" -0.303 0.032 -9.58 0 -0.367 -0.243
## 5 thin… ~ "a2" -0.266 0.031 -8.53 0 -0.328 -0.207
## 6 thin… ~~ "" 0.242 0.028 8.53 0 0.188 0.301
## 7 fake… ~~ "" 0.884 0.045 19.7 0 0.794 0.97
## 8 thin… ~~ "" 0.907 0.036 25.3 0 0.836 0.977
## 9 thin… ~~ "" 0.928 0.034 27.6 0 0.862 0.992
## 10 delu… ~~ "" 0.999 0 NA NA 0.999 0.999
## 11 indi… := "ind… 0.052 0.012 4.50 0 0.03 0.074
## 12 indi… := "ind… 0.038 0.01 3.65 0 0.018 0.058
## 13 tota… := "tot… 0.242 0.037 6.54 0 0.169 0.313
## # … with 3 more variables: std.lv <dbl>, std.all <dbl>, std.nox <dbl>
Missing data
Look for patterns of missingness
newz_md_tib %>%
dplyr::select(-id) %>%
mice::md.pattern(., rotate.names = TRUE)
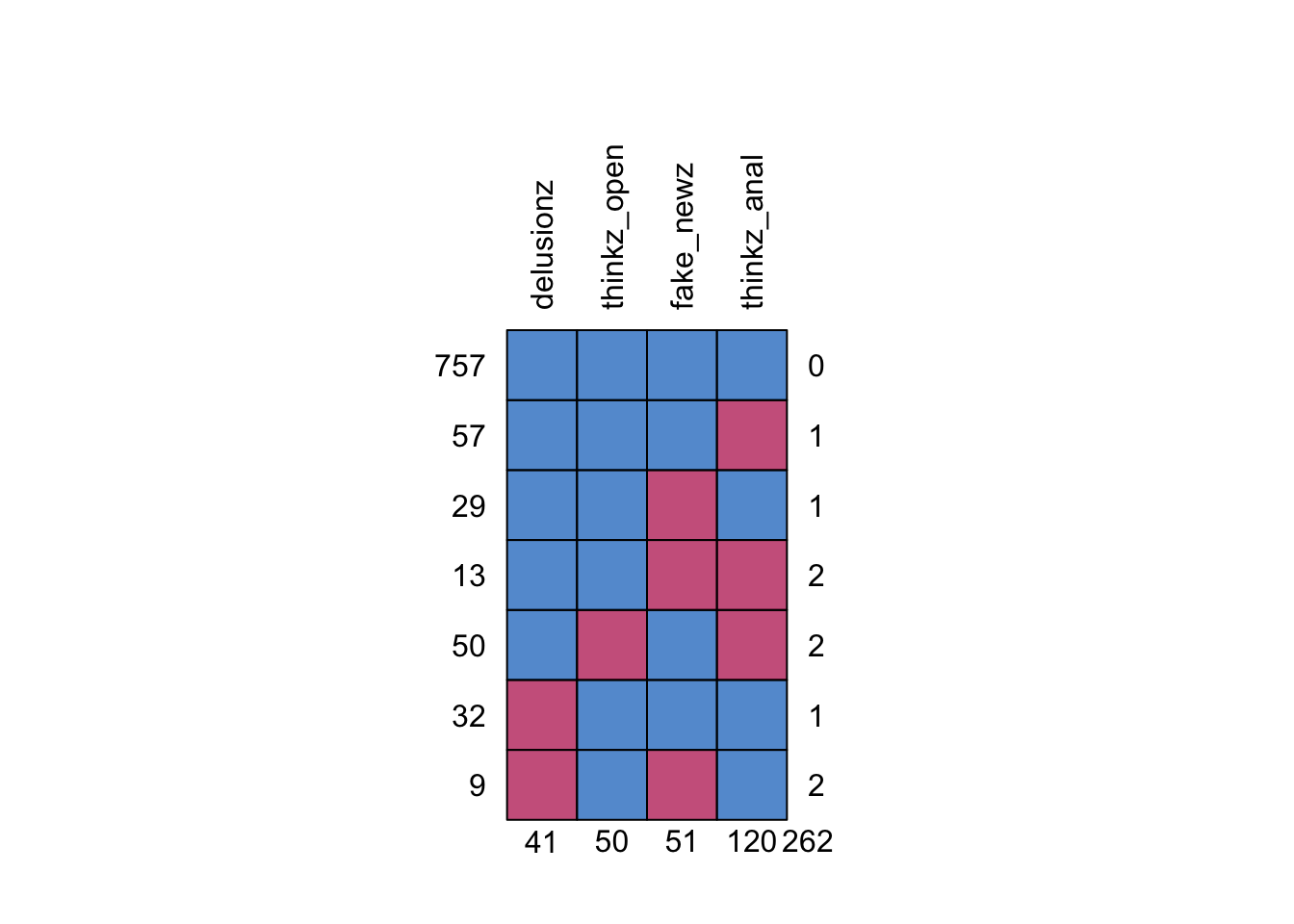
## delusionz thinkz_open fake_newz thinkz_anal
## 757 1 1 1 1 0
## 57 1 1 1 0 1
## 29 1 1 0 1 1
## 13 1 1 0 0 2
## 50 1 0 1 0 2
## 32 0 1 1 1 1
## 9 0 1 0 1 2
## 41 50 51 120 262
mice::ic(newz_md_tib)
## # A tibble: 190 x 5
## id thinkz_open fake_newz delusionz thinkz_anal
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 R_tEANRrR9dhnELbH -0.422 NA NA -1.86
## 2 R_2PqezczNjzLSZbG 1.36 NA -0.437 0.0633
## 3 R_UYHM5dj6jNuwnyV NA 1.08 -0.724 NA
## 4 R_rcKK0Qxk2wd2Az7 -1.11 NA 0.578 -0.417
## 5 R_2aXZNB6KOL9hfwn NA -1.38 0.813 NA
## 6 R_7OLg9j3w6u1lK25 NA 1.25 -1.22 NA
## 7 R_1dLg2gSsSQryr1i -1.24 -0.328 0.344 NA
## 8 R_3R32VYIZF29MaL8 0.810 NA -0.984 1.02
## 9 R_8uMt5tRNiusnHTr -1.11 NA 0.786 NA
## 10 R_3rM7LolOYmX4RkG 0.399 NA -0.828 0.543
## # … with 180 more rows
Listwise deletion (don’t do it!)
fake_lm <- lm(fake_newz ~ 0 + delusionz + thinkz_open + thinkz_anal, data = newz_md_tib)
broom::glance(fake_lm) %>%
dplyr::mutate(
dplyr::across(where(is.numeric), ~round(., 3))
)
## # A tibble: 1 x 12
## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 0.113 0.11 0.946 32.2 0 3 -1030. 2069. 2087.
## # … with 3 more variables: deviance <dbl>, df.residual <dbl>, nobs <dbl>
broom::tidy(fake_lm, conf.int = TRUE) %>%
dplyr::mutate(
dplyr::across(where(is.numeric), ~round(., 3))
)
## # A tibble: 3 x 7
## term estimate std.error statistic p.value conf.low conf.high
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 delusionz 0.164 0.036 4.51 0 0.093 0.236
## 2 thinkz_open -0.16 0.037 -4.29 0 -0.233 -0.087
## 3 thinkz_anal -0.134 0.037 -3.64 0 -0.207 -0.062
FIML
newz_lm <- 'fake_newz ~ delusionz + thinkz_open + thinkz_anal'
# If you want the intercept include 1
# newz_lm <- 'fake_newz ~ 1 + delusionz + thinkz_open + thinkz_anal'
newz_lm_fit <- lavaan::sem(newz_lm, data = newz_md_tib, missing = "fiml.x")
broom::tidy(newz_lm_fit, conf.int = TRUE) %>%
dplyr::mutate(
dplyr::across(where(is.numeric), ~round(., 3))
)
## # A tibble: 14 x 11
## term op estimate std.error statistic p.value conf.low conf.high std.lv
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 "fak… ~ 0.164 0.034 4.79 0 0.097 0.231 0.164
## 2 "fak… ~ -0.175 0.035 -5.02 0 -0.243 -0.107 -0.175
## 3 "fak… ~ -0.131 0.035 -3.70 0 -0.2 -0.061 -0.131
## 4 "fak… ~~ 0.873 0.041 21.1 0 0.792 0.955 0.873
## 5 "del… ~~ 0.986 0 NA NA 0.986 0.986 0.986
## 6 "del… ~~ -0.302 0 NA NA -0.302 -0.302 -0.302
## 7 "del… ~~ -0.26 0 NA NA -0.26 -0.26 -0.26
## 8 "thi… ~~ 1.01 0 NA NA 1.01 1.01 1.01
## 9 "thi… ~~ 0.327 0 NA NA 0.327 0.327 0.327
## 10 "thi… ~~ 0.996 0 NA NA 0.996 0.996 0.996
## 11 "fak… ~1 0.001 0.031 0.034 0.973 -0.06 0.062 0.001
## 12 "del… ~1 -0.001 0 NA NA -0.001 -0.001 -0.001
## 13 "thi… ~1 -0.005 0 NA NA -0.005 -0.005 -0.005
## 14 "thi… ~1 -0.013 0 NA NA -0.013 -0.013 -0.013
## # … with 2 more variables: std.all <dbl>, std.nox <dbl>
Multiple imputation
Find auxiliary variables
aux_vars <- mice::quickpred(newz_md_tib, exclude = "id", mincor = 0.1)
aux_vars
## id thinkz_open fake_newz delusionz thinkz_anal
## id 0 0 0 0 0
## thinkz_open 0 0 1 1 1
## fake_newz 0 1 0 1 1
## delusionz 0 1 1 0 1
## thinkz_anal 0 1 1 1 0
Imputation
newz_imps <- mice::mice(newz_md_tib, pred = aux_vars, method = "pmm", m = 20)
##
## iter imp variable
## 1 1 thinkz_open fake_newz delusionz thinkz_anal
## 1 2 thinkz_open fake_newz delusionz thinkz_anal
## 1 3 thinkz_open fake_newz delusionz thinkz_anal
## 1 4 thinkz_open fake_newz delusionz thinkz_anal
## 1 5 thinkz_open fake_newz delusionz thinkz_anal
## 1 6 thinkz_open fake_newz delusionz thinkz_anal
## 1 7 thinkz_open fake_newz delusionz thinkz_anal
## 1 8 thinkz_open fake_newz delusionz thinkz_anal
## 1 9 thinkz_open fake_newz delusionz thinkz_anal
## 1 10 thinkz_open fake_newz delusionz thinkz_anal
## 1 11 thinkz_open fake_newz delusionz thinkz_anal
## 1 12 thinkz_open fake_newz delusionz thinkz_anal
## 1 13 thinkz_open fake_newz delusionz thinkz_anal
## 1 14 thinkz_open fake_newz delusionz thinkz_anal
## 1 15 thinkz_open fake_newz delusionz thinkz_anal
## 1 16 thinkz_open fake_newz delusionz thinkz_anal
## 1 17 thinkz_open fake_newz delusionz thinkz_anal
## 1 18 thinkz_open fake_newz delusionz thinkz_anal
## 1 19 thinkz_open fake_newz delusionz thinkz_anal
## 1 20 thinkz_open fake_newz delusionz thinkz_anal
## 2 1 thinkz_open fake_newz delusionz thinkz_anal
## 2 2 thinkz_open fake_newz delusionz thinkz_anal
## 2 3 thinkz_open fake_newz delusionz thinkz_anal
## 2 4 thinkz_open fake_newz delusionz thinkz_anal
## 2 5 thinkz_open fake_newz delusionz thinkz_anal
## 2 6 thinkz_open fake_newz delusionz thinkz_anal
## 2 7 thinkz_open fake_newz delusionz thinkz_anal
## 2 8 thinkz_open fake_newz delusionz thinkz_anal
## 2 9 thinkz_open fake_newz delusionz thinkz_anal
## 2 10 thinkz_open fake_newz delusionz thinkz_anal
## 2 11 thinkz_open fake_newz delusionz thinkz_anal
## 2 12 thinkz_open fake_newz delusionz thinkz_anal
## 2 13 thinkz_open fake_newz delusionz thinkz_anal
## 2 14 thinkz_open fake_newz delusionz thinkz_anal
## 2 15 thinkz_open fake_newz delusionz thinkz_anal
## 2 16 thinkz_open fake_newz delusionz thinkz_anal
## 2 17 thinkz_open fake_newz delusionz thinkz_anal
## 2 18 thinkz_open fake_newz delusionz thinkz_anal
## 2 19 thinkz_open fake_newz delusionz thinkz_anal
## 2 20 thinkz_open fake_newz delusionz thinkz_anal
## 3 1 thinkz_open fake_newz delusionz thinkz_anal
## 3 2 thinkz_open fake_newz delusionz thinkz_anal
## 3 3 thinkz_open fake_newz delusionz thinkz_anal
## 3 4 thinkz_open fake_newz delusionz thinkz_anal
## 3 5 thinkz_open fake_newz delusionz thinkz_anal
## 3 6 thinkz_open fake_newz delusionz thinkz_anal
## 3 7 thinkz_open fake_newz delusionz thinkz_anal
## 3 8 thinkz_open fake_newz delusionz thinkz_anal
## 3 9 thinkz_open fake_newz delusionz thinkz_anal
## 3 10 thinkz_open fake_newz delusionz thinkz_anal
## 3 11 thinkz_open fake_newz delusionz thinkz_anal
## 3 12 thinkz_open fake_newz delusionz thinkz_anal
## 3 13 thinkz_open fake_newz delusionz thinkz_anal
## 3 14 thinkz_open fake_newz delusionz thinkz_anal
## 3 15 thinkz_open fake_newz delusionz thinkz_anal
## 3 16 thinkz_open fake_newz delusionz thinkz_anal
## 3 17 thinkz_open fake_newz delusionz thinkz_anal
## 3 18 thinkz_open fake_newz delusionz thinkz_anal
## 3 19 thinkz_open fake_newz delusionz thinkz_anal
## 3 20 thinkz_open fake_newz delusionz thinkz_anal
## 4 1 thinkz_open fake_newz delusionz thinkz_anal
## 4 2 thinkz_open fake_newz delusionz thinkz_anal
## 4 3 thinkz_open fake_newz delusionz thinkz_anal
## 4 4 thinkz_open fake_newz delusionz thinkz_anal
## 4 5 thinkz_open fake_newz delusionz thinkz_anal
## 4 6 thinkz_open fake_newz delusionz thinkz_anal
## 4 7 thinkz_open fake_newz delusionz thinkz_anal
## 4 8 thinkz_open fake_newz delusionz thinkz_anal
## 4 9 thinkz_open fake_newz delusionz thinkz_anal
## 4 10 thinkz_open fake_newz delusionz thinkz_anal
## 4 11 thinkz_open fake_newz delusionz thinkz_anal
## 4 12 thinkz_open fake_newz delusionz thinkz_anal
## 4 13 thinkz_open fake_newz delusionz thinkz_anal
## 4 14 thinkz_open fake_newz delusionz thinkz_anal
## 4 15 thinkz_open fake_newz delusionz thinkz_anal
## 4 16 thinkz_open fake_newz delusionz thinkz_anal
## 4 17 thinkz_open fake_newz delusionz thinkz_anal
## 4 18 thinkz_open fake_newz delusionz thinkz_anal
## 4 19 thinkz_open fake_newz delusionz thinkz_anal
## 4 20 thinkz_open fake_newz delusionz thinkz_anal
## 5 1 thinkz_open fake_newz delusionz thinkz_anal
## 5 2 thinkz_open fake_newz delusionz thinkz_anal
## 5 3 thinkz_open fake_newz delusionz thinkz_anal
## 5 4 thinkz_open fake_newz delusionz thinkz_anal
## 5 5 thinkz_open fake_newz delusionz thinkz_anal
## 5 6 thinkz_open fake_newz delusionz thinkz_anal
## 5 7 thinkz_open fake_newz delusionz thinkz_anal
## 5 8 thinkz_open fake_newz delusionz thinkz_anal
## 5 9 thinkz_open fake_newz delusionz thinkz_anal
## 5 10 thinkz_open fake_newz delusionz thinkz_anal
## 5 11 thinkz_open fake_newz delusionz thinkz_anal
## 5 12 thinkz_open fake_newz delusionz thinkz_anal
## 5 13 thinkz_open fake_newz delusionz thinkz_anal
## 5 14 thinkz_open fake_newz delusionz thinkz_anal
## 5 15 thinkz_open fake_newz delusionz thinkz_anal
## 5 16 thinkz_open fake_newz delusionz thinkz_anal
## 5 17 thinkz_open fake_newz delusionz thinkz_anal
## 5 18 thinkz_open fake_newz delusionz thinkz_anal
## 5 19 thinkz_open fake_newz delusionz thinkz_anal
## 5 20 thinkz_open fake_newz delusionz thinkz_anal
newz_mi_fit <- with(newz_imps, lm(fake_newz ~ 0 + delusionz + thinkz_open + thinkz_anal))
newz_pool <- mice::pool(newz_mi_fit)
summary(newz_pool, conf.int=TRUE)
## term estimate std.error statistic df p.value 2.5 %
## 1 delusionz 0.1629799 0.03489411 4.670700 488.0572 3.886229e-06 0.09441868
## 2 thinkz_open -0.1726764 0.03530186 -4.891425 453.2541 1.392987e-06 -0.24205201
## 3 thinkz_anal -0.1293757 0.03501683 -3.694673 472.2091 2.459457e-04 -0.19818381
## 97.5 %
## 1 0.23154113
## 2 -0.10330077
## 3 -0.06056765
summary(newz_pool, conf.int=TRUE) %>%
tibble::as_tibble() %>%
dplyr::mutate(
dplyr::across(where(is.numeric), ~round(., 3))
)
## # A tibble: 3 x 8
## term estimate std.error statistic df p.value `2.5 %` `97.5 %`
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 delusionz 0.163 0.035 4.67 488. 0 0.094 0.232
## 2 thinkz_open -0.173 0.035 -4.89 453. 0 -0.242 -0.103
## 3 thinkz_anal -0.129 0.035 -3.70 472. 0 -0.198 -0.061